props_to_image#
Values from the regionprops_3D function can be mapped back onto the original image.
import porespy as ps
import numpy as np
import matplotlib.pyplot as plt
import scipy.ndimage as spim
[17:46:05] ERROR PARDISO solver not installed, run `pip install pypardiso`. Otherwise, _workspace.py:56 simulations will be slow. Apple M chips not supported.
np.random.seed(7)
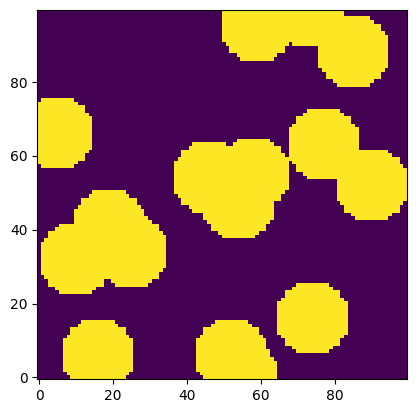
im = ~ps.generators.overlapping_spheres([100, 100], r=10, porosity=0.6)
plt.imshow(im, origin='lower', interpolation='none');
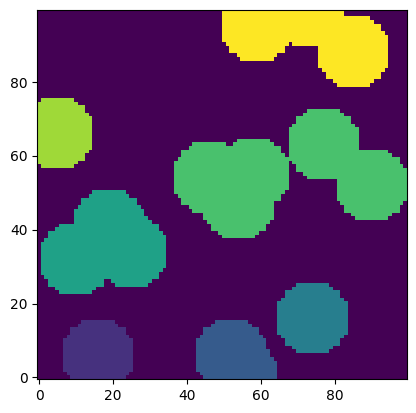
regions = spim.label(im)[0]
props = ps.metrics.regionprops_3D(regions)
plt.imshow(regions, origin='lower', interpolation='none');
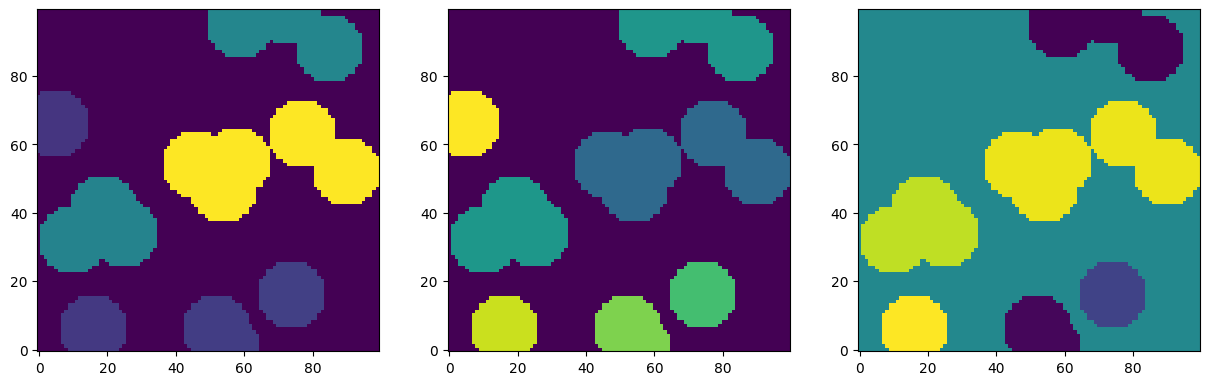
im1 = ps.metrics.prop_to_image(props, im.shape, 'convex_volume')
im2 = ps.metrics.prop_to_image(props, im.shape, 'sphericity')
im3 = ps.metrics.prop_to_image(props, im.shape, 'orientation')
fig, ax = plt.subplots(1, 3, figsize=[15, 5])
ax[0].imshow(im1, origin='lower', interpolation='none')
ax[1].imshow(im2, origin='lower', interpolation='none')
ax[2].imshow(im3, origin='lower', interpolation='none');