find_peaks#
Finds local peaks in the distance transform. This is need for watershed segmentations.
import numpy as np
import porespy as ps
import scipy.ndimage as spim
import matplotlib.pyplot as plt
from edt import edt
ps.visualization.set_mpl_style()
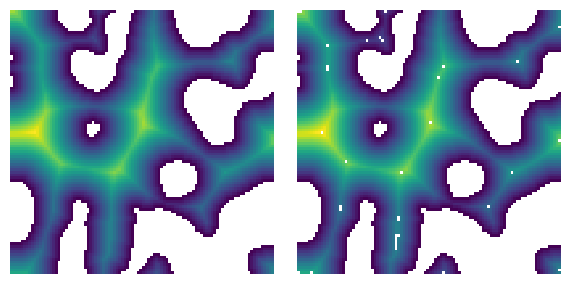
dt#
The distance transform must be provided
im = ps.generators.blobs(shape=[100, 100], blobiness=0.5, porosity=0.7, seed=0)
dt = edt(im)
fig, ax = plt.subplots(1, 2, figsize=[6, 3])
pk = ps.filters.find_peaks(dt=dt)
ax[0].imshow(dt / im)
ax[0].axis(False)
ax[1].imshow(dt / im / ~pk)
ax[1].axis(False);
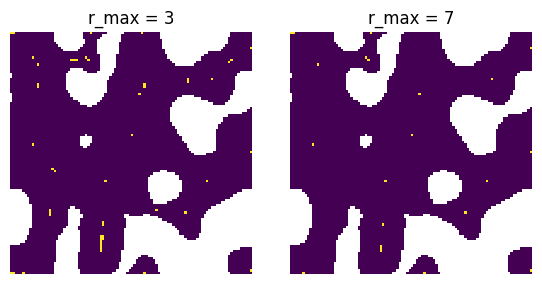
r_max#
The radius that should be searched for local maxima.
fig, ax = plt.subplots(1, 2, figsize=[6, 3])
r_max = 3
pk = ps.filters.find_peaks(dt=dt, r_max=r_max)
ax[0].imshow(pk / im)
ax[0].axis(False)
ax[0].set_title(f"r_max = {r_max}")
r_max = 7
pk = ps.filters.find_peaks(dt=dt, r_max=r_max)
ax[1].imshow(pk / im)
ax[1].axis(False)
ax[1].set_title(f"r_max = {r_max}");
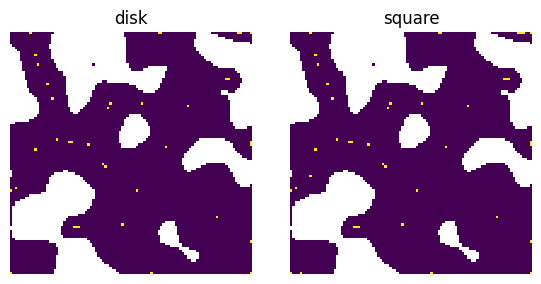
strel#
The structuring element to use.
im = ps.generators.blobs(shape=[100, 100], blobiness=0.5, porosity=0.7)
dt = edt(im)
from skimage.morphology import disk, square
fig, ax = plt.subplots(1, 2, figsize=[6, 3])
pk = ps.filters.find_peaks(dt=dt, strel=disk(3))
ax[0].imshow(pk / im)
ax[0].axis(False)
ax[0].set_title("disk")
pk = ps.filters.find_peaks(dt=dt, strel=square(5))
ax[1].imshow(pk / im)
ax[1].axis(False)
ax[1].set_title("square");
/tmp/ipykernel_21247/1099091553.py:13: FutureWarning: `square` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
pk = ps.filters.find_peaks(dt=dt, strel=square(5))