SNOW network extraction#
The SNOW algorithm, published in Physical Review E, uses a marker-based watershed segmentation algorithm to partition an image into regions belonging to each pore. The main contribution of the SNOW algorithm is to find a suitable set of initial markers in the image so that the watershed is not over-segmented. SNOW is an acronym for Sub-Network of an Over-segmented Watershed. This code works on both 2D and 3D images. In this example a 2D image will be segmented using the predefined snow function in PoreSpy.
Start by importing the necessary packages:
import matplotlib.pyplot as plt
import numpy as np
import openpnm as op
import porespy as ps
ps.visualization.set_mpl_style()
np.random.seed(10)
[19:37:41] WARNING PARDISO solver not installed on this platform. Simulations will be slow. _workspace.py:56
Generate an artificial 2D image for illustration purposes:
im = ps.generators.blobs(shape=[400, 400], porosity=0.6, blobiness=2)
fig, ax = plt.subplots(figsize=(4, 4))
ax.imshow(im);
SNOW is composed of a series of filters, but PoreSpy has a single function that applies all the necessary steps:
snow_output = ps.networks.snow2(im, voxel_size=1)
The snow function returns an object that has a network attribute. This is a dictionary that is suitable for loading into OpenPNM. The best way to get this into OpenPNM is to use the PoreSpy IO class. This splits the data into a network and a geometry:
pn = op.io.network_from_porespy(snow_output.network)
As can be seen by printing the network it contains quite a lot of geometric information:
print(pn)
══════════════════════════════════════════════════════════════════════════════
net : <openpnm.network.Network at 0x7f55e63afde0>
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
# Properties Valid Values
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
2 throat.conns 283 / 283
3 pore.coords 261 / 261
4 pore.region_label 261 / 261
5 pore.phase 261 / 261
6 throat.phases 283 / 283
7 pore.region_volume 261 / 261
8 pore.equivalent_diameter 261 / 261
9 pore.local_peak 261 / 261
10 pore.global_peak 261 / 261
11 pore.geometric_centroid 261 / 261
12 throat.global_peak 283 / 283
13 pore.inscribed_diameter 261 / 261
14 pore.extended_diameter 261 / 261
15 throat.inscribed_diameter 283 / 283
16 throat.total_length 283 / 283
17 throat.direct_length 283 / 283
18 throat.perimeter 283 / 283
19 pore.volume 261 / 261
20 pore.surface_area 261 / 261
21 throat.cross_sectional_area 283 / 283
22 throat.equivalent_diameter 283 / 283
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
# Labels Assigned Locations
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
2 pore.all 261
3 throat.all 283
4 pore.boundary 43
5 pore.xmin 12
6 pore.xmax 11
7 pore.ymin 10
8 pore.ymax 10
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
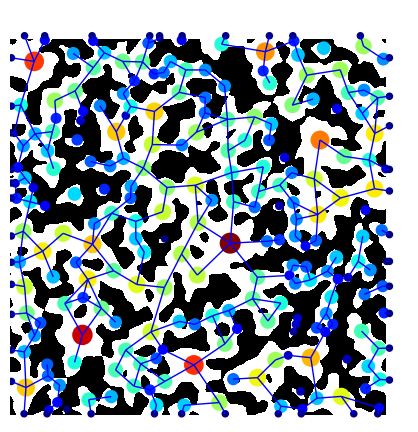
You can also overlay the network on the image natively in porespy. Note that you need to transpose the image using im.T, since imshow uses matrix representation, e.g. a (10, 20)-shaped array is shown as 10 pixels in the y-axis, and 20 pixels in the x-axis.
fig, ax = plt.subplots(figsize=[5, 5])
ax.imshow(im.T, cmap=plt.cm.bone)
op.visualization.plot_coordinates(
ax=fig,
network=pn,
size_by=pn["pore.inscribed_diameter"],
color_by=pn["pore.inscribed_diameter"],
markersize=200,
)
op.visualization.plot_connections(network=pn, ax=fig)
ax.axis("off");