MAGNET Network Extraction#
The MAGNET algorithm uses the traditional medial axis approach for extracting a network from an image. It makes use of the skeleton by locating pores as the junctions or endpoints of the skeleton.
import matplotlib.pyplot as plt
import numpy as np
import openpnm as op
import porespy as ps
ps.visualization.set_mpl_style()
np.random.seed(10)
[04:03:46] WARNING PARDISO solver not installed on this platform. Simulations will be slow. _workspace.py:56
Generate an Image#
im = ps.generators.blobs(shape=[400, 400], porosity=0.6, blobiness=2)
fig, ax = plt.subplots(figsize=(4, 4))
ax.imshow(im);
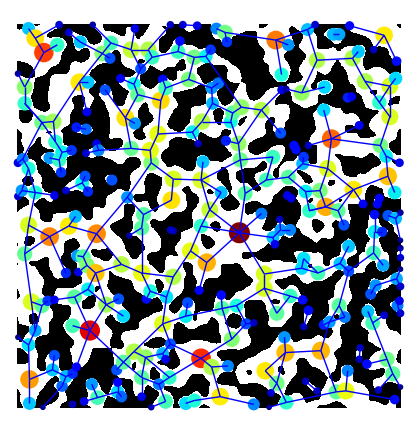
MAGNET uses a series of filters to extract a pore network but porespy has one function that does all this work for you!
magnet_output = ps.networks.magnet(im, voxel_size=1)
The magnet function returns an object that has a network attribute. This is a dictionary that is suitable for loading into OpenPNM. The best way to get this into OpenPNM is to use the PoreSpy IO class. This splits the data into a network and a geometry:
pn = op.io.network_from_porespy(magnet_output.network)
As can be seen by printing the network it contains quite a lot of geometric information:
print(pn)
══════════════════════════════════════════════════════════════════════════════
net : <openpnm.network.Network at 0x7ff623c8ac60>
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
# Properties Valid Values
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
2 throat.conns 327 / 327
3 pore.coords 301 / 301
4 throat.actual_length 327 / 327
5 throat.area 327 / 327
6 throat.max_diameter 327 / 327
7 throat.min_diameter 327 / 327
8 throat.avg_diameter 327 / 327
9 throat.inscribed_diameter 327 / 327
10 throat.integrated_diameter 327 / 327
11 pore.inscribed_diameter 301 / 301
12 pore.equivalent_diameter 301 / 301
13 pore.index 301 / 301
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
# Labels Assigned Locations
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
You can also overlay the network on the image natively in porespy. Note that you need to transpose the image using im.T, since imshow uses matrix representation, e.g. a (10, 20)-shaped array is shown as 10 pixels in the y-axis, and 20 pixels in the x-axis.
fig, ax = plt.subplots(figsize=[5, 5])
ax.imshow(im.T, cmap=plt.cm.bone)
op.visualization.plot_coordinates(
ax=fig,
network=pn,
size_by=pn["pore.inscribed_diameter"],
color_by=pn["pore.inscribed_diameter"],
markersize=200,
)
op.visualization.plot_connections(network=pn, ax=fig)
ax.axis("off");