Opening and Preparing Images#
The examples in the generators section have numerous examples on how to generate artificial images, but ultimately most people will be working with real images obtained from X-ray tomography or serial sectioning. This tutorial will cover a few different aspects about how to open these images for using a Jupyter notebook, or a Python workflow in general.
Let’s import the packages and functions that we’ll be needing:
import imageio.v2 as imageio
import numpy as np
from matplotlib.pyplot import subplots
from skimage.io import imread_collection
import porespy as ps
Opening a Folder of 2D tif Images#
Using ImageJ#
ImageJ is the ultimate image processing tool kit. It has many functions and filters for doing quantitative analysis, usually added via plug-ins (see note below), but it’s also very good at converting between file types and formats.
If you have a folder full of 2D images which are either microscope images obtained by serial sectioning or reconstructions of backprojections from X-ray tomography, then you’ll want to convert these into a single tif image. tif supports 3D images, so all the individual 2D images can be stacked together into a single file.
This can be done in ImageJ using “File/Import/Image Sequence” as shown in the screenshots below. Note that the images must be named with a numerical value indicating its order in the sequence, like “slice_001.tif”.

A dialog box will appear asking you about some of the details, which are usually correct by default:

Finally, you’ll want to save this new 3d tif image, which can be done with “save as”:

ImageJ and Fiji
ImageJ has a plugin architecture to add functionality. At some point it was realized that most users install numerous plugins, so Fiji was created, which is ImageJ with a large selection of the most popular plugins preinstalled. The name Fiji is actually a recursive acronym for “Fiji Is Just ImageJ”
Directly in Python#
If you’d rather not get manually involved, then it’s also possible to do this in Python. The scikit-image package comes with a handy funciton for just this purpose. It only takes a few minutes to get used to how it works:
seq = imread_collection("../../docs/_static/images/Sample 1/*.tif")
Now let’s inspect the result. We can see a list of files loaded in the files attribute:
seq.files
[]
And we can access the loaded images from the seq object as if it were a list:
fig, ax = subplots(figsize=[5, 5])
ax.imshow(seq[0]);
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
Cell In[4], line 2
1 fig, ax = subplots(figsize=[5, 5])
----> 2 ax.imshow(seq[0]);
File ~/work/porespy/porespy/.venv/lib/python3.13/site-packages/skimage/io/collection.py:306, in ImageCollection.__getitem__(self, n)
303 raise TypeError('slicing must be with an int or slice object')
305 if isinstance(n, int):
--> 306 n = self._check_imgnum(n)
307 idx = n % len(self.data)
309 if (self.conserve_memory and n != self._cached) or (self.data[idx] is None):
File ~/work/porespy/porespy/.venv/lib/python3.13/site-packages/skimage/io/collection.py:361, in ImageCollection._check_imgnum(self, n)
359 n = n % num
360 else:
--> 361 raise IndexError(f"There are only {num} images in the collection")
362 return n
IndexError: There are only 0 images in the collection
We can convert this list into a 3D image as follows:
im3d = np.zeros([*seq[0].shape, len(seq)])
for i, im in enumerate(seq):
im3d[..., i] = im
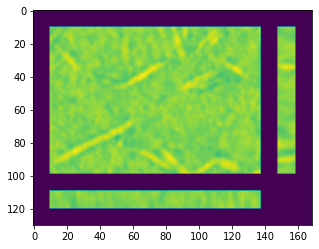
And now we can use porespy to visualize it:
fig, ax = subplots(figsize=[5, 5])
ax.imshow(ps.visualization.show_planes(im3d));
Convert to Binary#
In this tutorial we will not do anything to clean or denoise these images, as that deserves it’s own tutorial. Here we’ll just apply a threshold to convert a boolean with True indicating solid (the fibers above) and False indicating void.
First we need to look at the range of values in the image:
fig, ax = subplots(figsize=[5, 5])
ax.hist(im3d.flatten(), bins=25, edgecolor="k");
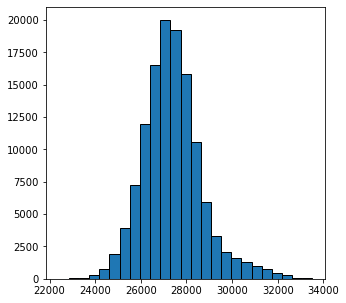
From the above histogram we can see that the values span from 22000 to 34000. We can just search for a value which does a decent job of segmenting the solid and void by trial and error:
fig, ax = subplots(figsize=[5, 5])
ax.imshow(im3d[..., 0] < 30000);

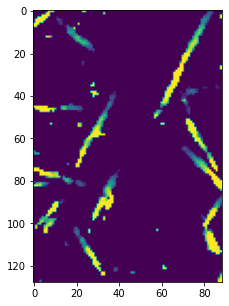
Because the image is noisy the result is not perfect. But let’s apply this threshold to the entire image now, then visualize the 3D stack:
im = im3d < 30000
fig, ax = subplots(figsize=[5, 5])
ax.imshow(ps.visualization.sem(im, axis=2));
Opening a 3D Stack#
Assuming that the 2D images have been turned into a 3D image, as described above, it is straightforward to load them:
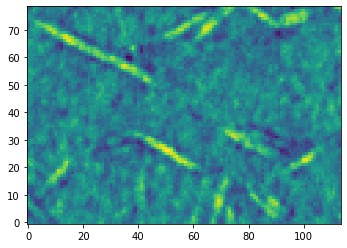
im2 = imageio.volread("../../docs/_static/images/Sample 1.tif")
ps.imshow(im2, axis=0);
Saving a 3D Image#
Once you have created a nice image that you want to save, perhaps for visualization in Paraview, you can also use the imageio package:
imageio.volsave('my image.tif', np.array(im, dtype=np.int8))
